SUNY Downstate and MetaCell launch a graphical tool to model, simulate and analyze brain neuronal circuits
CAMBRIDGE, Mass., Oct. 20, 2020 /PRNewswire/ —
Background
Unlocking the mysteries of the brain has become one of the major challenges for humanity in the 21st century, and it is likely to have a profound and transformative impact in both medicine and technology. However, despite the vastly expanding amount of neuroscience data being gathered, the development of approaches to interpret and gain meaningful insight from this data is lagging. Therefore, powerful new brain modeling tools that link the molecular, cellular and circuit scales, such as SUNY Downstate’s NetPyNE tool, are quickly gaining popularity amongst neuroscientists.
Introducing NetPyNE
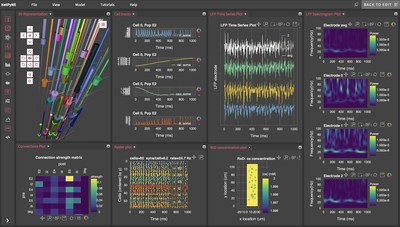
NetPyNE (Networks using Python and NEURON) is a project funded by the National Institute for Biomedical Imaging and Bioengineering at the NIH (U24 grant), the National Science Foundation (E-CAS grant), and the New York State Department of Health (SCIRB grant). NetPyNE provides a user-friendly, declarative language and a graphical interface to define the properties of molecules, neurons, and circuits. It also provides an easy way to validate the models against experimental data. The software is unique in that it allows students and scientists without programming expertise to build sophisticated brain models. NetPyNE also facilitates running parallel simulations on supercomputers and offers a variety of ways to visualize and analyze the resulting output, e.g. connectivity matrices, voltage traces, spike raster plots, local field potentials and information transfer measures.
The new NetPyNE GUI
The latest version of NetPyNE GUI (0.6.0), developed in partnership with MetaCell – a leading software company with deep domain expertise in computational neuroscience, molecular biology, data science, and enterprise-grade online software development – features numerous improvements that enable researchers to do everything they would normally do programmatically straight from a state-of-the-art and easy-to-use graphical interface. Through the web-based GUI, users can intuitively define their network models, visualize cells and networks in 3D, run simulations, and visualize and analyze data. The GUI includes an innovative, interactive Python console synchronized with the underlying Python-based model, making it incredibly easy to go back and forth from the coding to the GUI domain.
Resources
NetPyNE website | NetPyNE GUI | NetPyNE tutorials | NetPyNE GitHub
Salvador Durá-Bernal, Assistant Professor at SUNY Downstate Health Sciences University commented: “We’re excited to see the rapidly growing community of NetPyNE users. Our ultimate goal is to make this tool user-friendly enough such that the ‘non-computational community’ can also use it. The new GUI, developed with our partner MetaCell, is really impressive and will drive the continued success and adoption of the tool.”
Matteo Cantarelli, CTO and Co-Founder of MetaCell said: “MetaCell shares SUNY and the NetPyNE team’s vision of trying to decipher the brain’s neural coding mechanisms with far-reaching applications, including developing treatments for brain disorders. Researchers who take advantage of the new NetPyNE GUI will be able to run thousands of parallel simulations to explore how a range of parameters impact neuronal circuits.”
Photo – https://healthtechnologynet.com/wp-content/uploads/2020/10/MetaCell_SUNY_Downstate_GUI_NetPyNE.jpg
Contact details
MetaCell: www.metacell.us | info@metacell.us | @metacell | +1 (347) 878 3679 | +44 (0)7450 948685
View original content to download multimedia:http://www.prnewswire.com/news-releases/suny-downstate-and-metacell-launch-a-graphical-tool-to-model-simulate-and-analyze-brain-neuronal-circuits-301155221.html
SOURCE MetaCell LLC
